Ongoing Research Projects
Research Summary
My research is driven by a passion for understanding the complexities of life through the lens of genomics and computational biology. Whether through developing innovative software tools such as PG-SUI, advancing species delimitation techniques with SuperDeli, or exploring the evolutionary dynamics of hybrid zones, my goal is to contribute to the scientific community's ability to study and conserve the natural world in an era of rapid environmental change and habitat destruction.

GeoGenIE: Geographic-Genetic Inference Engine
GeoGenIE (Geographic-Genetic Inference Engine) is an advanced software tool designed to use deep learning for predicting geographic localities (latitude and longitude) based on genetic SNP data inputs. Built on PyTorch, GeoGenIE is specifically optimized to account for geographic sampling bias and the limited number of SNPs typically found in GT-seq panels. This software is a powerful resource for researchers who need to infer geographic origins from genetic data, providing a robust framework for handling complex datasets.
GeoGenIE not only excels in prediction accuracy compared to other existing software, but also offers a wealth of metrics and visualizations that allow users to thoroughly assess model performance. The software is highly user-friendly, ensuring that even those with limited experience in deep learning can effectively utilize its capabilities. GeoGenIE is also highly efficient, with optional parallelized bootstrapping across multiple CPU cores to speed up analysis. Users have extensive flexibility in modifying model parameters, visualization options, and preprocessing settings, all while adhering to standard best practices. GeoGenIE leverages vectorized Python modules such as numpy, scipy, scikit-learn, and pandas, ensuring both speed and reliability in processing.
SNPio: Object-Oriented Python API for Popultion Genomic Data Filtering, Visualization, and File Conversion
SNPio is a cutting-edge software project aimed at creating a user-friendly, object-oriented Python API designed for reading, filtering, and converting various standard genomic data file formats. In the complex field of genomics, researchers often face challenges when dealing with diverse data formats and the need for efficient data manipulation tools. SNPio addresses these challenges by providing a streamlined interface that simplifies the management of genomic datasets, enabling users to focus on analysis rather than data handling.
This software is particularly valuable because it brings together a wide range of functionalities within a single framework, making it easier for researchers to process and analyze genomic data. With SNPio, tasks like filtering large SNP datasets, converting between formats (e.g., VCF, STRUCTURE, PHYLIP), and integrating data from different sources are made straightforward and accessible, even to those with limited programming experience. Ongoing development efforts include expanding the API’s capabilities to support additional file formats, enhancing performance, calculating summary statistics ad visualization, and integrating SNPio with popular bioinformatics tools and pipelines.

Genomic SNP Imputation using AI (PG-SUI)
One of my key research projects is the development of PG-SUI (Population Genomics - Supervised and Unsupervised Imputation), an AI-based framework for imputing missing single nucleotide polymorphism (SNP) data in genomic datasets. Missing data is a common issue in population genomics, often leading to biased results and reduced statistical power. PG-SUI addresses this challenge by employing both supervised and unsupervised machine learning models in a user-friendly manner to accurately predict missing SNPs based on non-linear patterns observed in the existing data.
This project is particularly significant because it not only enhances the accuracy of population genomic studies but also enables researchers to utilize incomplete datasets - a commonplace issue in population genetics - more effectively. By reducing the need for extensive data collection, PG-SUI accelerates research timelines and lowers costs, making genomic studies more accessible to a broader range of scientists. Ongoing work involves refining the existing models to handle a wider variety of data types, adding new models, and integrating the framework into commonly used bioinformatics pipelines.

Species Delimitation using Machine Learning (SuperDeli)
Another current focus of my research is the application of machine learning techniques to species delimitation—a process crucial for understanding biodiversity, conservation, and evolutionary biology. Traditional methods of species delimitation often rely on subjective interpretations of morphological and genetic data, leading to inconsistent results. By incorporating machine learning, my approach provides a more objective and reproducible framework for defining species boundaries.
The machine learning models I develop for species delimitation take into account both genetic and ecological data, allowing for a comprehensive analysis that considers multiple factors influencing species differentiation. This approach is particularly useful in cases where species exhibit subtle morphological differences or where hybridization complicates the identification of distinct species. My work in this area aims to create tools that are not only scientifically rigorous but also user-friendly, so they can be widely adopted by researchers in various fields of biology.
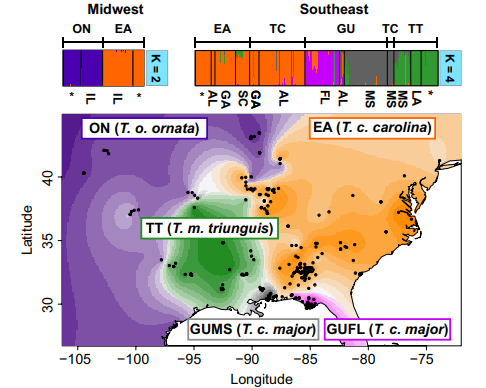
Evolutionary Dynamics of Hybrid Zones
Hybrid zones—regions where distinct species interbreed—are natural laboratories for studying evolutionary processes. My research in this area focuses on understanding the genetic and environmental factors that drive hybridization and the formation of new species. By combining field data with genomic analyses, I investigate how hybrid zones contribute to biodiversity and the evolution of new species. This research has significant implications for conservation biology, particularly in understanding how human activities influence hybridization and species integrity.
Collaborations and Future Directions
My work is highly collaborative, involving partnerships with researchers in ecology, evolutionary biology, and computational and data science. These collaborations allow me to apply cutting-edge computational methods to a wide range of biological questions, from the dynamics of disease resistance in wildlife populations to the genomic underpinnings of speciation. Looking forward, I aim to expand my research to include more interdisciplinary projects that integrate ecological modeling, climate change predictions, and conservation strategies.
In the future, I plan to continue advancing the use of AI and machine learning in genomics, with a particular focus on making these tools more accessible to non-experts. This involves not only developing more intuitive software and user interfaces but also providing training and resources to help researchers apply these techniques to their own work.